Super-Resolution Reveals the Basis of Interchromosomal Linkages
Super-resolution microscopy has allowed scientists at the Stowers Institute for Medical Research to identify what causes interchromosomal linkages. Their findings cited our c-Myc lentivirus and were published in the Journal of Cell Biology.
The composition of interchromosomal linkages
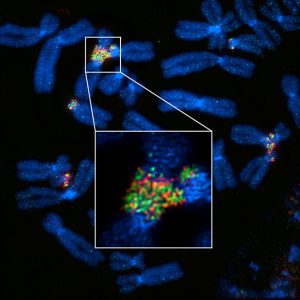
Structural illumination super-resolution microscopy (SIM) images of mitotic chromosomes from human cells, showing rDNA-linked chromosomes. rDNA was labeled with rDNA probe and UBF antibody (green and red). Both rDNA and UBF form filamentous connections between chromosomes. Credit: Gerton Lab, Stowers Institute for Medical Research.
Interchromosomal linkages were discovered by researchers in the 1960s while studying the organization of the human genome; however, due to the resolution limitations, these linkages were difficult to visualize. They were able to observe that the associations occurred between the short ends of the five different acrocentric chromosomes, in which the centromere is positioned near the end of the chromosome, but the basis of these linkages remained unknown.
The Stowers Institute researchers were able to reveal that the chromosomes were linked by a shared ribosomal DNA (rDNA) sequence located on the short arms of the chromosomes. Their research also exposed the positive correlation between transcription and the frequency of interchromosomal connections at specific genomic regions. While examining human mammary epithelial cells (HMECs), they observed heightened frequency of the associations in the HMEC line that was transduced with c-Myc. C-Myc binds directly to the rDNA accelerating transcription of the 45s gene.
Importance of resolution
The team’s research also uncovered the dependence upon topoisomerase II for the formation as well as the resolution of these rDNA linkages. Topoisomerase II regulates DNA supercoiling during transcription by cutting and rejoining DNA strands, relieving torsional stress and increasing the rate of transcription. The scientists also speculated that the topoisomerase could entangle the rDNA sequences from chromosomes and create the linkages. Despite its role in the formation of the chromosomal connections, it was also determined that it is responsible for the resolution of the connections during the metaphase-to-anaphase transition and therefore does not contribute to chromosomal missegregation and genomic instability.
These new discoveries can provide insights into Robertsonian translocations, fusions between chromosomes that contain rDNA, which can lead to infertility and trisomies such as Down syndrome. Thanks to this research, we are one step closer to fully understanding the complexity of genomic organization.
The c-Myc lentivirus used in the study is a premade lentivirus containing c-Myc, a regulatory gene that codes for a transcription factor believed to be responsible for 15% of all gene expression. You can view all our premade lentiviral vectors here, or check out other unique reagents for chromosome research:
- DNA-RNA Hybrid [S9.6] Antibody from National Institute of Allergy and Infectious Diseases/NIH
- 3T3 Translocation Cell Lines from National Cancer Institute/NIH
- DNA Damage/Repair Transformed MEF Cell Lines from National Institute of Environmental Health Sciences/NIH
- MarathonRT Reverse Transcriptase from Yale University
DNA Polymerase Eta Antibody from Icahn School Of Medicine at Mount Sinai


