Testing for antibiotic-resistant bacteria in 30 minutes
Antibiotics are incredibly important due to their ability to prevent and treat bacterial infections, such as strep throat, sinus infections and pneumonia. However, the misuse of antibiotics has reduced their efficacy, with an increasing number of bacteria species becoming antibiotic-resistant. According to the World Health Organization, “Antibiotic resistance is one of the biggest threats to global health, food security, and development today.”
Researchers at Caltech have now developed a test that can identify antibiotic-resistant bacteria in as little as 30 minutes. Their study, published this month in Science Translational Medicine, could help healthcare professionals select the best antibiotic for treating a given infection.
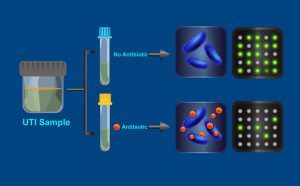
How the antibiotic resistance test works. Credit: Caltech.
The research team focused on urinary tract infections (UTIs). More than 50% of women contract a UTI in their lifetimes, resulting in eight million doctor visits and one million ER visits in the United States annually. Their goal was to develop a test for antibiotic-resistant bacteria that could be completed during a visit to the doctor’s office.
To complete the test, a patient with a UTI gives a urine sample. The sample is divided into two parts, the first of which is exposed to an antibiotic for 15 minutes. The second part is incubated without exposure to antibiotics. Then, each sample’s bacteria are lysed to release their cellular contents and a detection chemistry technique is used to image and individually count specific DNA markers. The entire process can be completed in under 30 minutes.
The test is based on the idea that bacteria typically cannot replicate their DNA as well in an antibiotic solution, resulting in fewer of the DNA markers. If the bacteria are resistant to the antibiotic, however, the DNA replication will continue as normal. In that scenario, the test will show similar numbers of DNA markers in both the treated and untreated solutions. In the study, the researchers tested this method on 54 patients with UTIs. The results showed a 95% match with those obtained using the standard, currently accepted test, which takes two days to return results.
The ability to rapidly identify antibiotic-resistant bacteria is important for healthcare professionals making treatment decisions. When doctors treat bacterial infections, they often skip over the antibiotics to which bacteria are more likely to be resistant, going straight to stronger, second-line antibiotics. Though this increases the chance that the individual’s treatment will be effective, it also makes it more likely that bacteria will begin to develop resistance to stronger antibiotics as well.
“Therapies are driven by guidelines developed by organizations like the World Health Organization or the Centers for Disease Control and Prevention without knowing what the patient actually has, because the tests are so slow,” senior author Dr. Rustem Ismagilov said in a press release. “We can change the world with a rapid test like this. We can change the way antibiotics are prescribed.”
Up next, the research team will be testing this method on other types of infectious bacteria. In addition, they hope to adjust the method so that tests could also be run on blood samples.
Do you work in this area of science? Check out our available bacteriology reagents to see if any could help accelerate your research. You also might be interested in past blog posts covering antibiotic research:


