Development of Photocleavable Alpha-Factor for Yeast
Laurie L. Parker, PhD, Purdue University — Published: March 20, 2013
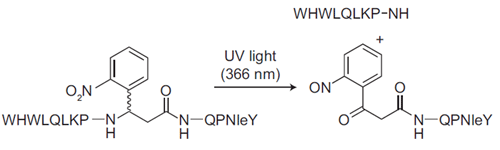
Diagram of Photocleavable Alpha-Factor for Yeast.This photocleavable alpha-factor analog incorporates a UV-cleavable linker into the backbone, which enables photolysis of the peptide upon exposure to UV light. This mimics Bar1 protease cleavage and cell cycle release.
As a synthetic chemist moving into a new field and learning the language of biology during my postdoctoral training with Steve Kron and Steve Kent, I wanted to bring my molecular skills to the table in a way that would connect to my new environment. I was receiving state of the art training in peptide chemistry in the Kent group, and spent a large proportion of my time surrounded by yeast geneticists in the Kron group. One day Steve Kron and I were tossing ideas back and forth and he brought up the α-factor peptide—Steve had always thought it would be cool to make an engineered version in which degradation could be controlled. It clicked that this was the perfect combination: structural design, peptide synthesis and yeast biology—all of the elements coming together!
Lucky for me, Dr. Fred Naider and Dr. Jeffrey Becker had done a lot of work describing how the structure of the α-factor pheromone peptide contributed to its function (Naider, F. and Becker, J. Peptides, 2004). They had found that replacing a central glycine in the peptide with D-alanine actually enhanced the peptide’s activity—presumably by stabilizing the bent conformation needed for activity. That this central glycine could be replaced with bulkier amino acids (as long as the stereochemistry was controlled) was a good sign—it suggested that replacing it with a cleavable linker of some kind would be possible.
We chose 3-amino-3-(2-nitrophenyl)propionic acid as a linker because it was small and cheap—only the racemate was available, but since the product peptides are diastereomers, they were resolvable during purification. Initially I had to call these diastereomers the “right” peak and the “left” peak, since all I knew about them was that one eluted later than the other during reverse phase HPLC. I tested them using the classical α-factor arrest “shmoo” assay (looking for morphological changes in MATa yeast cells in the presence of the peptide) and found that the diastereomer in the “right” peak was active but the one in the “left” peak was not.
To test release from arrest after photocleavage of the peptide, I needed a more quantitative assay—it’s possible to count shmoos vs. buds as a way to quantify arrest or release, but trying to pick out the shmoo vs. bud “needles” in the field of view “haystack” made my head hurt! So I found a higher-throughput, less subjective method: flow cytometry for DNA content, which helped me establish that cells could be arrested with this analog and released using UV light. Based on Naider and Becker’s work, I had a hunch that the diastereomer in this “right” peak contained the D isomer of the photolinker. I worked with Dr. Josh Kurutz, then director of the biological NMR facility at the University of Chicago, to analyze the stereochemistry and indeed, when he assigned the structure for each of the peptides I had isolated (right and left peaks), he found evidence that the active material was the peptide containing the D-photolinker. All in all a very neat wrap-up to my first project as a chemical biologist.
Learn more about Dr. Parker’s Photocleavable Alpha Factor for Yeast »
Tags:
Alpha-factor, cell cycle, cell cycle arrest, MATa cells, mating-specific genes, Bar1 protease, Photocleavable Alpha-Factor for Yeast
References:
- Parker LL, Kurutz JW, Kent SBH, and Kron SJ. Control of the Yeast Cell Cycle with a Photocleavable Alpha-Factor Analogue. Angew. Chem., Int. Ed. 2006, 38, 6322–6325.



