New insights into ubiquitin regulation
Research from the University of Würzburg is providing new insights into ubiquitin regulation. In a recent study, published in the journal eLife, researchers determined the atomic structure of a ubiquitin ligase implicated in tumor formation, revealing a dimerization mechanism that may be exploited for developing new cancer therapies. The paper cited Kerafast’s LOBSTR E. coli Expression Strain, created by Dr. Thomas U. Schwartz at the Massachusetts Institute of Technology for one-step downstream purification of recombinant polyhistidine-tagged proteins.
Ubiquitous ubiquitin
Ubiquitin is a small protein, found in almost all tissues of humans and other eukaryotic organisms, with significant regulatory influence. First discovered in 1975, ubiquitin is added to target proteins via a process called ubiquitination. Ubiquitination can have a wide range of effects on the substrate protein, causing degradation or changes in activity or cellular location.
One group of enzymes involved in the ubiquitination process are called ubiquitin ligases. In the current study, the research team looked specifically at a ubiquitin ligase called HUWE1. HUWE1 is an extremely large protein, with almost 4,400 amino acids, and it has been shown to play a key role in tumorigenesis. However, details about how the ligase’s activity is regulated, as well as its three-dimensional structure, have remained mostly unknown.
It takes two
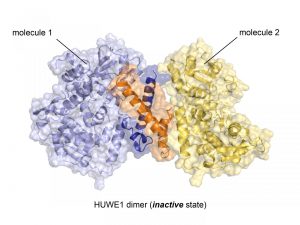
Crystal structure of the ubiquitin ligase HUWE1 in the newly discovered inactive state. The two molecules that form the dimer are represented as cartoons and surfaces. The region that mediates dimer formation is highlighted (orange, dark blue). Credit: Sonja Lorenz.
The University of Würzburg researchers used X-ray crystallography to determine the atomic structure of the C-terminal part of HUWE1. They found a surprising new feature of the protein: two HUWE1 molecules can pair up to form a dimer complex, which in turn inhibits enzymatic activity.
The research team then explored how this dimerization is regulated so that each cell maintains the proper balance of inactive dimers and single, active molecules. They determined that the tumor suppressor protein p14ARF can bind to HUWE1, causing a shift toward the inactive dimer state. The p14ARF protein is often non-present in cancer cells, and this new study offers the first explanation of how p14ARF works to inhibit HUWE1 – a finding that may provide new avenues for cancer therapy.
“The effects of p14ARF on the structure and activity of HUWE1 are extremely exciting,” said senior author Dr. Sonja Lorenz in a press release. “They open up a range of possibilities to manipulate HUWE1 activity that we are following up on.”
Related reagents
Do you work in this area of research? Check out the LOBSTR E. coli Expression Strain, which the investigators in the study used to express HUWE1 fragments. You also might be interested in one of our reagents for studying ubiquitin, including:
- MultiDsk Ubiquitin Binding Protein Reagent from Cancer Research Technology
- U-tag SUMO Trapping Protein from College of William & Mary
- E3 Ubiquitin-Protein Ligase RNF5 Antibody from Mount Sinai School of Medicine
- Ubiquitin-Conjugating Enzyme E2 R1 (CDC34) Monobody from University of Illinois at Chicago


