MarathonRT Ultra-Processive Reverse Transcriptase in Action
Reverse transcriptases (RTs), enzymes that turn RNA to the more stable DNA, are crucial research tools, but leading RTs face limitations like low processivity, reactivity at temperatures that degrade RNA, and the inability to unwind RNA structures. MarathonRT Reverse Transcriptase, available through Kerafast, an Absolute Biotech brand, helps solve these problems.
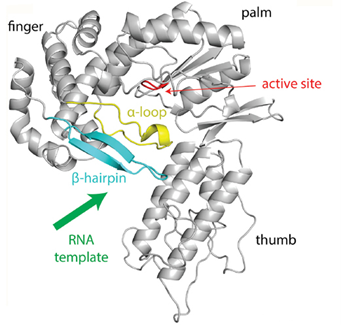
MarathonRT is a group II intron RT purified from Eubacterium rectale and characterized with novel features, like the alpha loop and the beta hairpin, which can melt template structures within RNA without falling off, even at ambient temperatures. Compared to other high-performance RTs like Superscript IV (SSIV), MarathonRT has superior processivity in transcripts at least 10 kb in length, while retaining comparable low error frequency (Zhao & Pyle, 2018). MarathonRT’s ability to copy long RNA transcripts end-to-end allows for the detection of variation in entire transcriptomes. MarathonRT has been at the forefront of new research in areas like oncology and virology. We have collected just a few of the recent developments in which MarathonRT has been involved.
Recent publications using MarathonRT
ALL-tRNAseq enables robust tRNA profiling in tissue samples
We got to see this reagent in action in the 2023 publication from Amsterdam UMC researchers who used MarathonRT and RNA demethylation to develop a robust way to profile tRNA. Alterations in tRNA populations can affect mRNA translational efficiency during cancer progression, so the accurate measuring of tRNA in patient samples is crucial to getting an accurate oncogenic profile of the cancer patients.
Nano-DMS-MaP allows isoform-specific RNA structure determination
This 2023 Nature Methods article highlights MarathonRT’s ability to generate long cDNAs in the presence of RNA modifications and confirms that MarathonRT nearly exclusively generates single nucleotide substitutions at positions of DMS modification. These qualities aided researchers in developing nanopore dimethylsulfate mutational profiling (Nano-DMS-MaP), a method that provides isoform-resolved structural information of similar RNA molecules. Methods like Nano-DMS-MaP can help understanding of complex viral machinery for pathogens like HIV, as well as deepen understanding of the structures that control gene regulation.
Systematic detection of tertiary structural modules in large RNAs and RNP interfaces by Tb-seq
Another methodology to identify RNA structures was published in Nature Communications in 2023. In compact 3D RNA structures, the negatively-charged RNA backbones are often in tight quarters, so cations are recruited to stabilize the structure. Researchers used MarathonRT to develop a sequencing approach to identifying terbium(III) (Tb3+) cleavage points in long RNA strands, thereby revealing dense 3D RNA structures within the strand. MarathonRT can navigate complex structures at the magnitude of multi-kilobases to help researchers identify and characterize the RNA structures that dictate gene expression.
Conclusions
MarathonRT’s ability to copy transcripts of at least 10 kb in length without falling off RNA structures earned it the “Marathon” moniker and continues to enable researchers to understand facets of gene expression, viral pathogenesis, and more previously limited by existing RTs. MarathonRT is available through Kerafast, an Absolute Biotech brand devoted to making unique laboratory reagents accessible to the wider scientific community. Other reagents available through Absolute Biotech brands for DNA and RNA research include:
- SCICONS antibodies against double-stranded RNA, available through Nordic-MUbio/Exalpha
- DNA-RNA Hybrid Antibody from National Institute of Allergy and Infectious Diseases/NIH, available through Kerafast
- DNA/RNA Research Antibodies from Absolute Antibody
If you have questions about MarathonRT or any of our products, please contact us. We are happy to help!


